Example 3: Longitudinal Sections
This example demonstrates how longitudinal sections data read by PT3S can be accessed and used to create plots with Matplotlib.
PT3S Release
[1]:
#pip install PT3S -U --no-deps
Necessary packages for this Example
When running this example for the first time on your machine, please execute the cell below. Afterward, you may need to restart the kernel (using the ‘fast-forward’ button).
[2]:
pip install -q bokeh
Note: you may need to restart the kernel to use updated packages.
Imports
[3]:
import os
import logging
import pandas as pd
import datetime
import numpy as np
import subprocess
import matplotlib
import matplotlib.pyplot as plt
import matplotlib.dates as mdates
import matplotlib.gridspec as gridspec
import matplotlib.ticker as ticker
import matplotlib.colors as mcolors
from matplotlib.pyplot import Polygon
from matplotlib.ticker import FuncFormatter
from matplotlib.dates import DateFormatter, MinuteLocator
import matplotlib.ticker as ticker
from bokeh.plotting import figure, show
from bokeh.io import output_notebook
from bokeh.models import CustomJS, ColumnDataSource, CheckboxGroup, LinearAxis, Range1d
from bokeh.layouts import column
try:
from PT3S import dxAndMxHelperFcts
except:
import dxAndMxHelperFcts
try:
from PT3S import Rm
except:
import Rm
[4]:
import importlib
from importlib import resources
[5]:
#importlib.reload(dxAndMxHelperFcts)
Logging
[6]:
logger = logging.getLogger()
if not logger.handlers:
logFileName = r"Example3.log"
loglevel = logging.DEBUG
logging.basicConfig(
filename=logFileName,
filemode='w',
level=loglevel,
format="%(asctime)s ; %(name)-60s ; %(levelname)-7s ; %(message)s"
)
fileHandler = logging.FileHandler(logFileName)
logger.addHandler(fileHandler)
consoleHandler = logging.StreamHandler()
consoleHandler.setFormatter(logging.Formatter("%(levelname)-7s ; %(message)s"))
consoleHandler.setLevel(logging.INFO)
logger.addHandler(consoleHandler)
Read Model and Results
[7]:
dbFilename="Example3"
dbFile = resources.files("PT3S").joinpath("Examples", f"{dbFilename}.db3")
[8]:
m=dxAndMxHelperFcts.readDxAndMx(dbFile=dbFile
,preventPklDump=True
,maxRecords=-1
,mxsVecsResults2MxDfVecAggs=[7,13,19,-1]
#,SirCalcExePath=r"C:\3S\SIR 3S\SirCalc-90-14-02-12_Potsdam.fix1_x64\SirCalc.exe"
)
INFO ; Dx.__init__: dbFile (abspath): c:\users\aUserName\3s\pt3s\PT3S\Examples\Example3.db3 exists readable ...
INFO ; PT3S.dxAndMxHelperFcts.readDxAndMx:
+..\PT3S\Examples\Example3.db3 is newer than
+..\PT3S\Examples\WDExample3\B1\V0\BZ1\M-1-0-1.1.MX1:
+SIR 3S' dbFile is newer than SIR 3S' mx1File
+in this case the results are maybe dated or (worse) incompatible to the model
INFO ; PT3S.dxAndMxHelperFcts.readDxAndMx:
+..\PT3S\Examples\WDExample3\B1\V0\BZ1\M-1-0-1.XML is newer than
+..\PT3S\Examples\WDExample3\B1\V0\BZ1\M-1-0-1.1.MX1:
+SirCalc's xmlFile is newer than SIR 3S' mx1File
+in this case the results are maybe dated or (worse) incompatible to the model
INFO ; PT3S.dxAndMxHelperFcts.readDxAndMx: Model is being recalculated using C:\\3S\SIR 3S Entwicklung\SirCalc-90-15-02-03_Quebec_x64\SirCalc.exe
INFO ; Mx.setResultsToMxsFile: Mxs: ..\PT3S\Examples\WDExample3\B1\V0\BZ1\M-1-0-1.1.MXS reading ...
INFO ; dxWithMx.__init__: Example3: processing dx and mx ...
Longitudinal Sections: V3_AGSN
[9]:
m.V3_AGSN.head()
[9]:
| Pos | pk | tk | LFDNR | NAME | XL | compNr | nextNODE | OBJTYPE | OBJID | ... | RHO_n | mlc_n | ('STAT', 'mlc', Timestamp('2023-02-12 23:00:00'), Timestamp('2023-02-12 23:00:00'))_n | ('TIME', 'mlc', Timestamp('2023-02-12 23:00:00'), Timestamp('2023-02-12 23:00:00'))_n | ('TMIN', 'mlc', Timestamp('2023-02-12 23:00:00'), Timestamp('2023-02-13 23:00:00'))_n | ('TMAX', 'mlc', Timestamp('2023-02-12 23:00:00'), Timestamp('2023-02-13 23:00:00'))_n | ('TIME', 'mlc', Timestamp('2023-02-13 06:00:00'), Timestamp('2023-02-13 06:00:00'))_n | ('TIME', 'mlc', Timestamp('2023-02-13 12:00:00'), Timestamp('2023-02-13 12:00:00'))_n | ('TIME', 'mlc', Timestamp('2023-02-13 18:00:00'), Timestamp('2023-02-13 18:00:00'))_n | ('TIME', 'mlc', Timestamp('2023-02-13 23:00:00'), Timestamp('2023-02-13 23:00:00'))_n | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | -1 | 5755933101669454049 | 5755933101669454049 | 1.0 | Längsschnitt | 0 | 1 | V-E0 | ROHR | 5691533564979419761 | ... | 965.700012 | 592.989116 | 592.964582 | 592.964582 | 592.96461 | 603.808833 | 602.036334 | 599.220665 | 602.192192 | 593.485427 |
| 0 | 0 | 5755933101669454049 | 5755933101669454049 | 1.0 | Längsschnitt | 0 | 1 | V-K1683S | ROHR | 5691533564979419761 | ... | 965.701172 | 592.964582 | 592.964582 | 592.964582 | 592.96461 | 603.808833 | 602.036334 | 599.220665 | 602.192192 | 593.485427 |
| 1 | 1 | 5755933101669454049 | 5755933101669454049 | 1.0 | Längsschnitt | 0 | 1 | V-K1693S | ROHR | 5048873293262650113 | ... | 965.702148 | 592.944601 | 592.944601 | 592.944601 | 592.944655 | 603.733968 | 601.97052 | 599.16915 | 602.125588 | 593.462842 |
| 2 | 2 | 5755933101669454049 | 5755933101669454049 | 1.0 | Längsschnitt | 0 | 1 | V-K2163S | ROHR | 5715081934973525403 | ... | 965.702637 | 592.934593 | 592.934593 | 592.934593 | 592.934659 | 603.696467 | 601.93755 | 599.14334 | 602.09222 | 593.451528 |
| 3 | 3 | 5755933101669454049 | 5755933101669454049 | 1.0 | Längsschnitt | 0 | 1 | V-K2043S | ROHR | 5413647981880727734 | ... | 965.703735 | 592.911548 | 592.911548 | 592.911548 | 592.911641 | 603.610202 | 601.861708 | 599.083971 | 602.015467 | 593.425478 |
5 rows × 85 columns
Plot Section No. 1
Define Axes
[10]:
def fyPH(ax,offset=0):
ax.spines["left"].set_position(("outward", offset))
ax.set_ylabel('PH Druck in bar')
#ax.set_ylim(1,6)
#ax.set_yticks(sorted(np.append(np.linspace(1,6,11),[])))
ax.yaxis.set_ticks_position('left')
ax.yaxis.set_label_position('left')
def fymlc(ax,offset=60):
ax.spines["left"].set_position(("outward", offset))
ax.set_ylabel('mlc Druckhöhe in mlc')
#ax.set_ylim(1,6)
#ax.set_yticks(sorted(np.append(np.linspace(1,6,11),[])))
ax.yaxis.set_ticks_position('left')
ax.yaxis.set_label_position('left')
def fybarBzg(ax,offset=120):
ax.spines["left"].set_position(("outward", offset))
ax.set_ylabel('H Druck in barBzg')
#ax.set_ylim(1,6)
#ax.set_yticks(sorted(np.append(np.linspace(1,6,11),[])))
ax.yaxis.set_ticks_position('left')
ax.yaxis.set_label_position('left')
def fyM(ax,offset=180):
Rm.pltLDSHelperY(ax)
ax.spines["left"].set_position(("outward",offset))
ax.set_ylabel('QM Massenstrom in t/h')
#ax.set_ylim(500,550)
#ax.set_yticks(sorted(np.append(np.linspace(500,550,11),[])))
ax.yaxis.set_ticks_position('left')
ax.yaxis.set_label_position('left')
def fyT(ax,offset=240):
Rm.pltLDSHelperY(ax)
ax.spines["left"].set_position(("outward",offset))
ax.set_ylabel('T Tempertatur in °C')
ax.set_ylim(55,95)
#ax.set_yticks(sorted(np.append(np.linspace(0,95,11),[])))
ax.yaxis.set_ticks_position('left')
ax.yaxis.set_label_position('left')
Plotfunction
[11]:
def plot(dfAGSN=pd.DataFrame()
,dfAGSNRL=pd.DataFrame()
,PHCol='PH_n'
,mlcCol='mlc_n'
,zKoorCol='ZKOR_n'
,barBzgCol='H_n'
,QMCol='QM'
,TCol='T_n'
,xCol='LSum'
):
fig, ax0 = plt.subplots(figsize=Rm.DINA3q)
ax0.set_yticks(np.linspace(0, 10, 21))
ax0.yaxis.set_ticklabels([])
ax0.grid()
#PH
ax1 = ax0.twinx()
fyPH(ax1)
PH_SL=ax1.plot(dfAGSN[xCol], dfAGSN[PHCol], color='red', label='PH SL',ls='dotted')
PH_RL=ax1.plot(dfAGSNRL[xCol], dfAGSNRL[PHCol], color='blue', label='PH RL',ls='dotted')
#mlc
ax11 = ax0.twinx()
fymlc(ax11)
mlc_SL=ax11.plot(dfAGSN[xCol], dfAGSN[mlcCol], color='red', label='mlc SL')
mlc_RL=ax11.plot(dfAGSNRL[xCol], dfAGSNRL[mlcCol], color='blue', label='mlc RL')
z=ax11.plot(dfAGSN[xCol], dfAGSN[zKoorCol], color='black', label='z',ls='dashed',alpha=.5)
#barBZG
ax12 = ax0.twinx()
fybarBzg(ax12)
barB_SL=ax12.plot(dfAGSN[xCol], dfAGSN[barBzgCol], color='red', label='H SL',ls='dashdot')
barB_RL=ax12.plot(dfAGSNRL[xCol], dfAGSNRL[barBzgCol], color='blue', label='H RL',ls='dashdot')
#M
ax2 = ax0.twinx()
fyM(ax2)
QM_SL=ax2.step(dfAGSN[xCol], dfAGSN[QMCol]*dfAGSN['direction'], color='orange', label='M SL')
QM_RL=ax2.step(dfAGSNRL[xCol], dfAGSNRL[QMCol]*dfAGSNRL['direction'], color='cyan', label='M RL',ls='--')
#T
ax3 = ax0.twinx()
fyT(ax3)
T_SL=ax3.plot(dfAGSN[xCol], dfAGSN[TCol], color='pink', label='T SL')
T_RL=ax3.plot(dfAGSNRL[xCol], dfAGSNRL[TCol], color='lavender', label='T RL')
ax0.set_title('Longitudinal Section for '+dbFilename)
# added these three lines
lns = PH_SL+ PH_RL + mlc_SL+ mlc_RL + barB_SL+ barB_RL+ QM_SL+ QM_RL + T_SL+ T_RL + z
labs = [l.get_label() for l in lns]
ax0.legend(lns, labs)#, loc=0)
plt.show()
Plot without vector data
[12]:
dfAGSN=m.V3_AGSN[
(m.V3_AGSN['LFDNR']==1)
&
(m.V3_AGSN['XL']==1)
]
[13]:
dfAGSNRL=m.V3_AGSN[
(m.V3_AGSN['LFDNR']==1)
&
(m.V3_AGSN['XL']==2)
]
[14]:
plot(dfAGSN,dfAGSNRL)
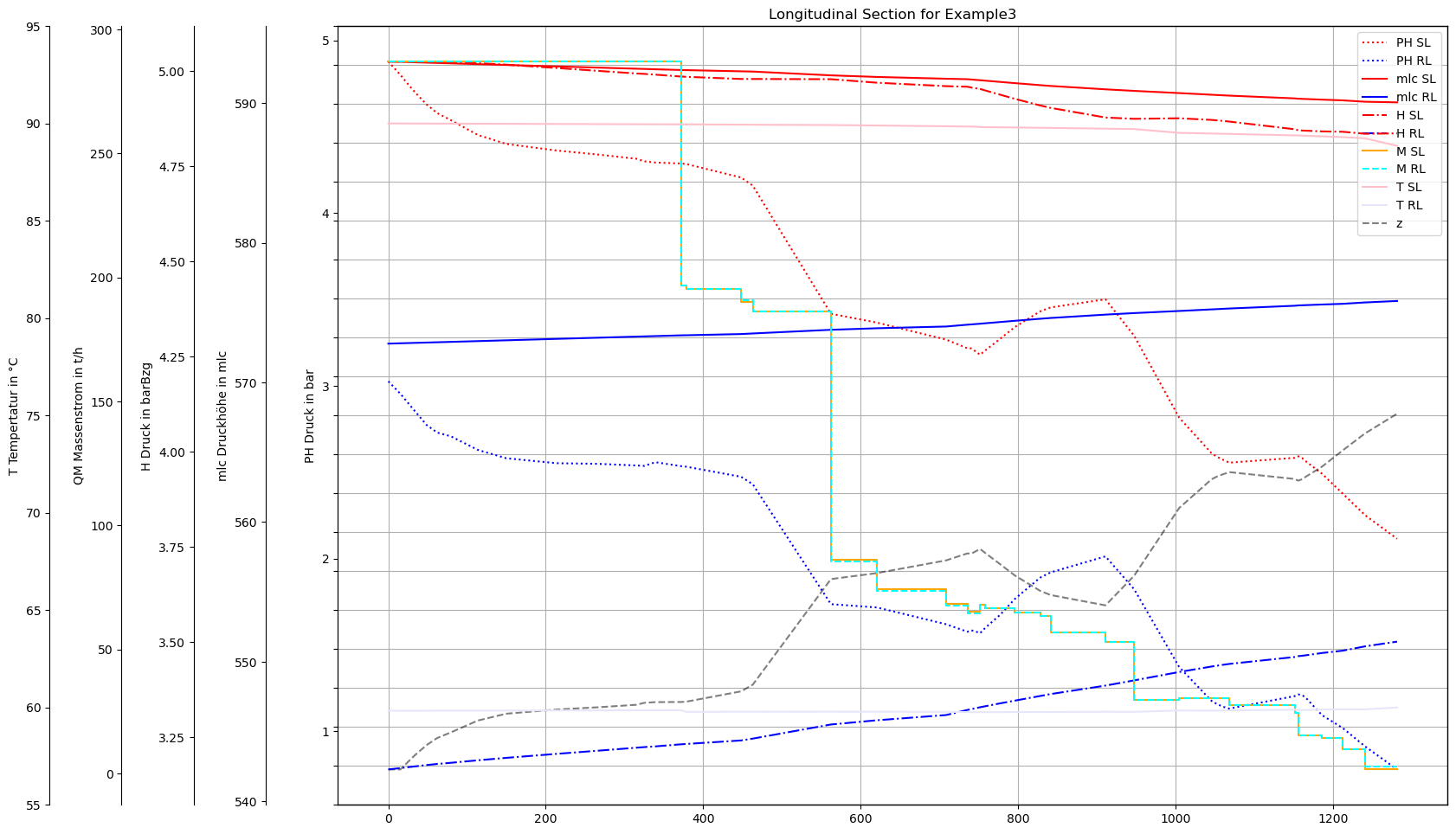
Plot with vector data (with pipe interior points)
[15]:
dfAGSNVec=m.V3_AGSNVEC[
(m.V3_AGSNVEC['LFDNR']==1)
&
(m.V3_AGSNVEC['XL']==1)
]
[16]:
dfAGSNVecRL=m.V3_AGSNVEC[
(m.V3_AGSNVEC['LFDNR']==1)
&
(m.V3_AGSNVEC['XL']==2)
]
[17]:
t0rVec=pd.Timestamp(m.mx.df.index[0].strftime('%Y-%m-%d %X'))#.%f'))
[18]:
manPVEC=('STAT',
'manPVEC',
t0rVec,
t0rVec)
[19]:
manPVEC
[19]:
('STAT',
'manPVEC',
Timestamp('2023-02-12 23:00:00'),
Timestamp('2023-02-12 23:00:00'))
[20]:
# for convenient colnames in celloutput
dfAGSNVec['manPVEC']=dfAGSNVec[manPVEC]
dfAGSNVecRL['manPVEC']=dfAGSNVecRL[manPVEC]
[21]:
mlcPVEC=('STAT',
'mlcPVEC',
t0rVec,
t0rVec)
[22]:
# for convenient colnames in celloutput
dfAGSNVec['mlcPVEC']=dfAGSNVec[mlcPVEC]
dfAGSNVecRL['mlcPVEC']=dfAGSNVecRL[mlcPVEC]
[23]:
QMVEC=('STAT',
'QMVEC',
t0rVec,
t0rVec)
[24]:
QMVEC=('STAT',
'QMVEC',
t0rVec,
t0rVec)
[25]:
# for convenient colnames in celloutput
dfAGSNVec['QMVEC']=dfAGSNVec[QMVEC]
dfAGSNVecRL['QMVEC']=dfAGSNVecRL[QMVEC]
[26]:
TVEC=('STAT',
'ROHR~*~*~*~TVEC',
t0rVec,
t0rVec)
[27]:
# for convenient colnames in celloutput
dfAGSNVec['TVEC']=dfAGSNVec[TVEC]
dfAGSNVecRL['TVEC']=dfAGSNVecRL[TVEC]
[28]:
plot(dfAGSNVec,dfAGSNVecRL
,PHCol=manPVEC
,mlcCol=mlcPVEC
,zKoorCol='ZVEC'
,barBzgCol='H_n'
,QMCol=QMVEC
,TCol=TVEC
,xCol='LSum'
)
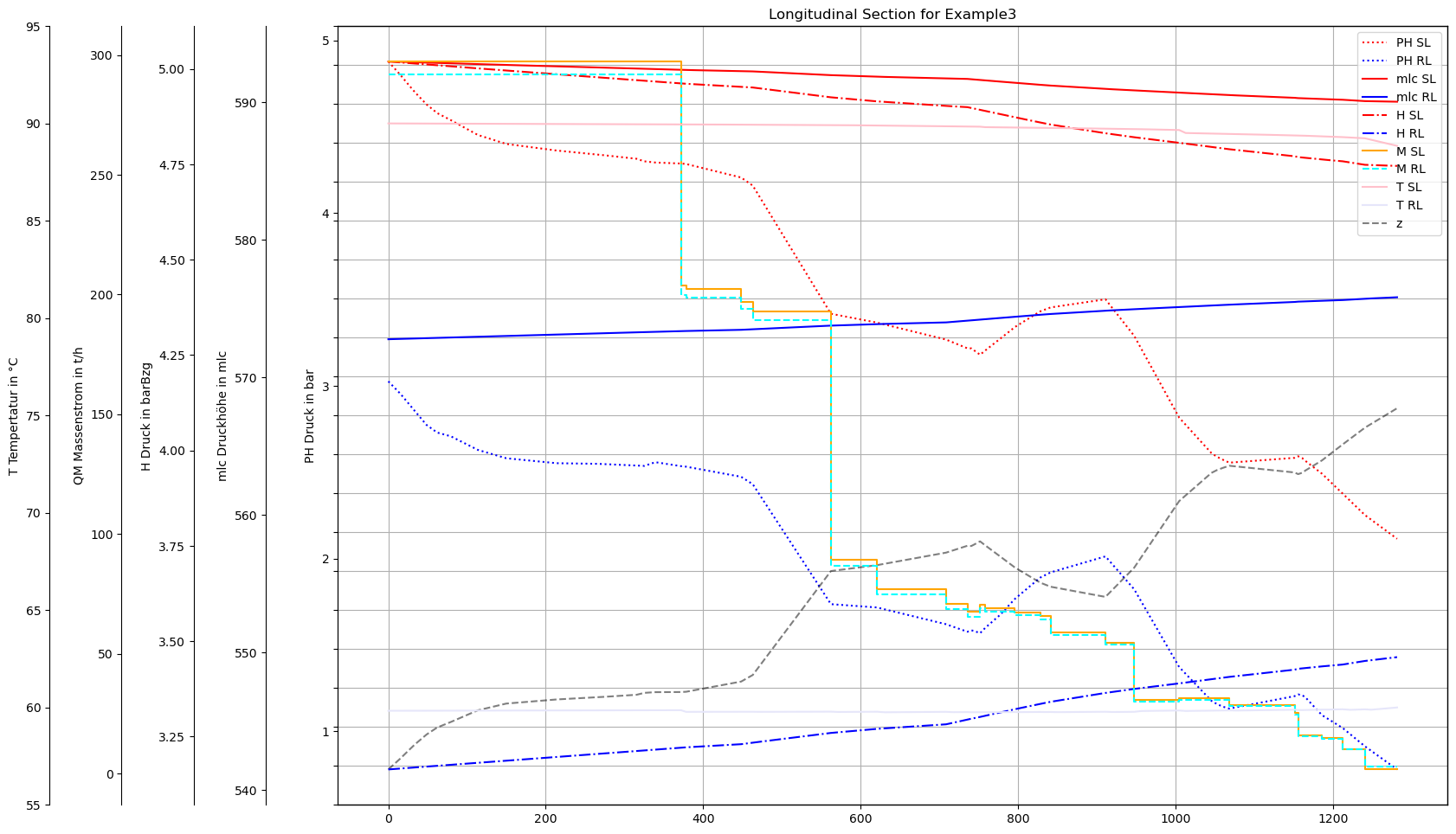
Plot different timesteps with Bokeh
[29]:
def bokeh_plot(df, x_col, y_cols, y2_cols, y3_cols):
output_notebook()
p = figure(width=1366, height=768, title="Longitudinal Section for "+dbFilename+" at different timestamps", x_axis_label=x_col, y_axis_label="PH Druck in bar", tools="pan,wheel_zoom,box_zoom,reset", y_range=Range1d(start=0, end=6))
# Define line styles and colors
line_styles = ['solid', 'dashed', 'dotted']
colors = ['blue', 'red', 'green']
lines = []
# lines for y
for i, y_col in enumerate(y_cols):
line = p.line(df[x_col], df[y_col], legend_label=f"{y_col} (06:00)" if i == 0 else f"{y_col} (12:00)" if i == 1 else f"{y_col} (18:00)", line_width=2, line_dash=line_styles[i % len(line_styles)], color=colors[0])
lines.append(line)
#second y-axis
p.extra_y_ranges = {"y2": Range1d(start=0, end=600)}
p.add_layout(LinearAxis(y_range_name="y2", axis_label="QM Massenstrom in t/h"), 'left')
# lines for y2
for i, y2_col in enumerate(y2_cols):
line = p.line(df[x_col], df[y2_col], legend_label=f"{y2_col} (06:00)" if i == 0 else f"{y2_col} (12:00)" if i == 1 else f"{y2_col} (18:00)", line_width=2, line_dash=line_styles[i % len(line_styles)], color=colors[1], y_range_name="y2")
lines.append(line)
#third y-axis
p.extra_y_ranges["y3"] = Range1d(start=590, end=610)
p.add_layout(LinearAxis(y_range_name="y3", axis_label="mlc in m"), 'left')
# lines for y3
for i, y3_col in enumerate(y3_cols):
line = p.line(df[x_col], df[y3_col], legend_label=f"{y3_col} (06:00)" if i == 0 else f"{y3_col} (12:00)" if i == 1 else f"{y3_col} (18:00)", line_width=2, line_dash=line_styles[i % len(line_styles)], color=colors[2], y_range_name="y3")
lines.append(line)
# Create a CheckboxGroup
labels = [f"{y_col}" for i, y_col in enumerate(y_cols + y2_cols + y3_cols)]
checkbox = CheckboxGroup(labels=labels, active=list(range(len(labels))))
# CustomJS to toggle visibility
checkbox.js_on_change('active', CustomJS(args=dict(lines=lines), code="""
for (let i = 0; i < lines.length; i++) {
lines[i].visible = this.active.includes(i);
}
"""))
show(column(p, checkbox))
[30]:
bokeh_plot(dfAGSNVec, 'LSum',['PH_n_1', 'PH_n_2', 'PH_n_3'], ['QM_1', 'QM_2', 'QM_3'], ['mlc_n_1', 'mlc_n_2', 'mlc_n_3'])